Main Page: Difference between revisions
Jump to navigation
Jump to search
(Added new plugin Cavitomix) |
mNo edit summary |
||
| Line 30: | Line 30: | ||
|- | |- | ||
! New Plugin | ! New Plugin | ||
| [[CavitOmiX|CavitOmiX]] calculate [https://innophore.com Catalophore™ | | [[CavitOmiX|CavitOmiX]] calculate [https://innophore.com Catalophore™ cavities], predict protein structures with [https://www.nvidia.com/en-us/gpu-cloud/bionemo OpenFold by NVIDIA-BioNeMo], [https://ai.facebook.com/blog/protein-folding-esmfold-metagenomics/ ESMFold] and retrieve [https://www.deepmind.com/research/highlighted-research/alphafold Alphafold] models | ||
|- | |- | ||
! Official Release | ! Official Release | ||
Revision as of 07:08, 24 December 2022
| The community-run support site for the PyMOL molecular viewer. |
| To request a new account, email SBGrid at: accounts (@) sbgrid dot org |
| Tutorials | Table of Contents | Commands |
| Script Library | Plugins | FAQ |
| Gallery | Covers | PyMOL Cheat Sheet (PDF) | Getting Help |
|
|
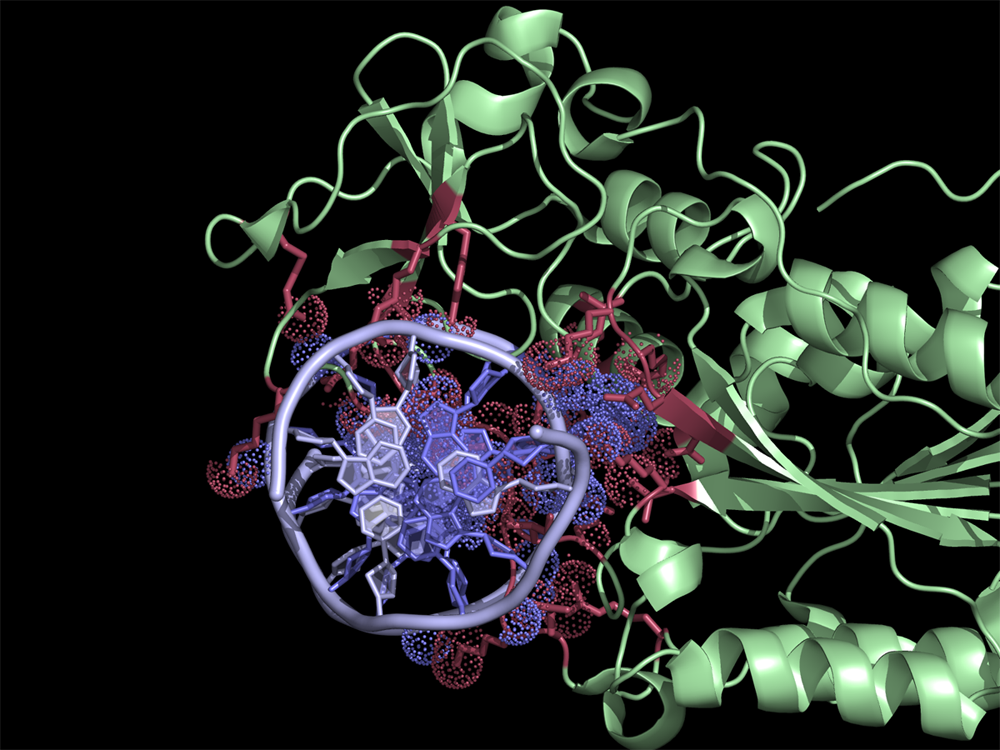
 A Random PyMOL-generated Cover. See Covers.
| ||||||||||||||||||||||||||||||||