Split selection
Jump to navigation
Jump to search
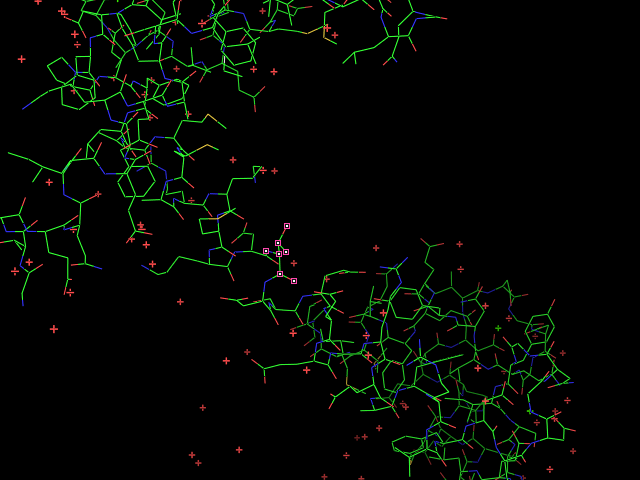
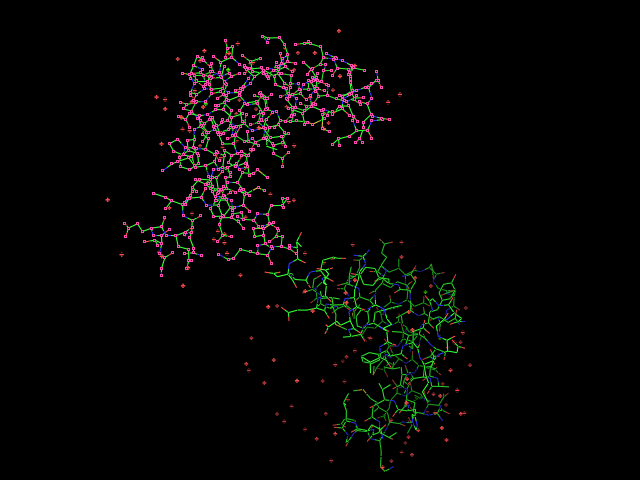
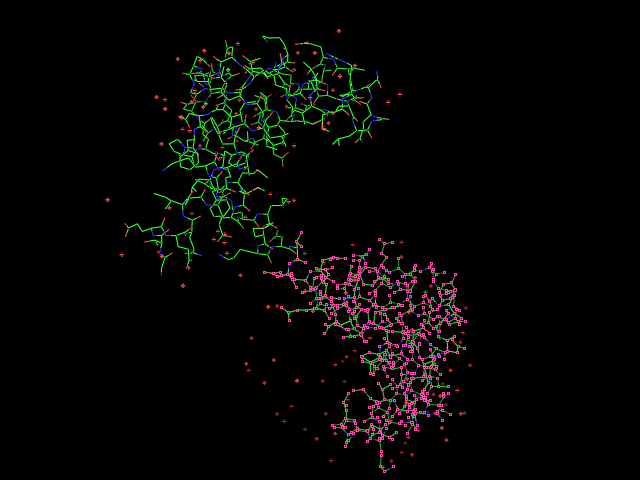
Given an initial selection, Split_selection will create two new selections. One, called 'lo,' will have all the atoms with IDs lower than your input atom (actually input residue's alpha carbon); and the second selection is 'hi,' which contains the atoms with IDs higher than the initial (residue's alpha carbon) atom.
The Code
import pymol
from pymol import cmd
def get_index_list(s):
"""
Given an atom selection, return the list of atom IDs in this selection
"""
return map(lambda x: x.index, cmd.get_model(s).atom)
def get_single_index(s):
"""
Get the ID of the first alpha carbon in the selection, S
"""
# assume CA
return get_index_list( "n. CA and (br. %s)" % s)[0]
def split_selection(s):
"""
PARAMS
s
An atom selection.
RETURNS
None
SIDE EFFECTS
Creates two new selections, called lo and hi. Lo will have all atoms in the same molecule
with atom IDs less than the alpha carbon ID in S. Hi will have all the atoms in the same
molecule with IDs greater than the atom ID of the alpha carbon in S.
EXAMPLE
run /path/to/this/script/split_selection.py
fetch 1cll
select i. 79
split_selection (sele)
# now look at the 'hi' and 'lo' selections.
AUTHOR: Jason Vertrees, 2010.
"""
l = get_index_list("bm. " + s)
m = min(l)
M = max(l)
# assume using alpha carbons
selected_index = get_single_index( "n. CA and (br. sele)" )
low_sel_name = cmd.get_unused_name("lo")
hi_sel_name = cmd.get_unused_name("hi")
cmd.select(low_sel_name, "ID %d-%d" % (m,selected_index-1))
cmd.select(hi_sel_name, "ID %d-%d" % (selected_index+1,M))
cmd.extend("split_selection", split_selection)