PDB plugin
PDB PyMOL plugin
PDBe’s PyMOL plugin provides an easy way to visualise PDB data and annotations in PyMOL. This includes displaying individual molecules, Pfam, SCOP and CATH domains and viewing geometric validation for PDB entries based on PDBe's API.
Installation
The PDB PyMOL plugin works on PyMOL 1.6 or later
PyMOL 1.6
Download the plugin from the following URL:
http://www.ebi.ac.uk/pdbe/pdb-component-library/pymol_plugin/PDB_plugin.py
PyMOL 1.7 or later
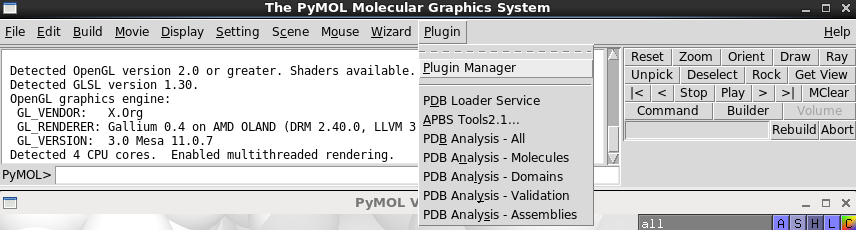
Open the “plugin manager” Enter the following url in the “Install from PyMOL Wiki or URL
http://www.ebi.ac.uk/pdbe/pdb-component-library/pymol_plugin/PDB_plugin.py
This will add the following items to the Plugins menu:
PDB Analysis - All
PDB Analysis - Molecules
PDB Analysis - Domains
PDB Analysis - Validation
PDB Analysis - Assemblies
Screenshot of the menu
To use each component click the menu item in the Plugins menu and then enter and PDB ID into the dialogue box.
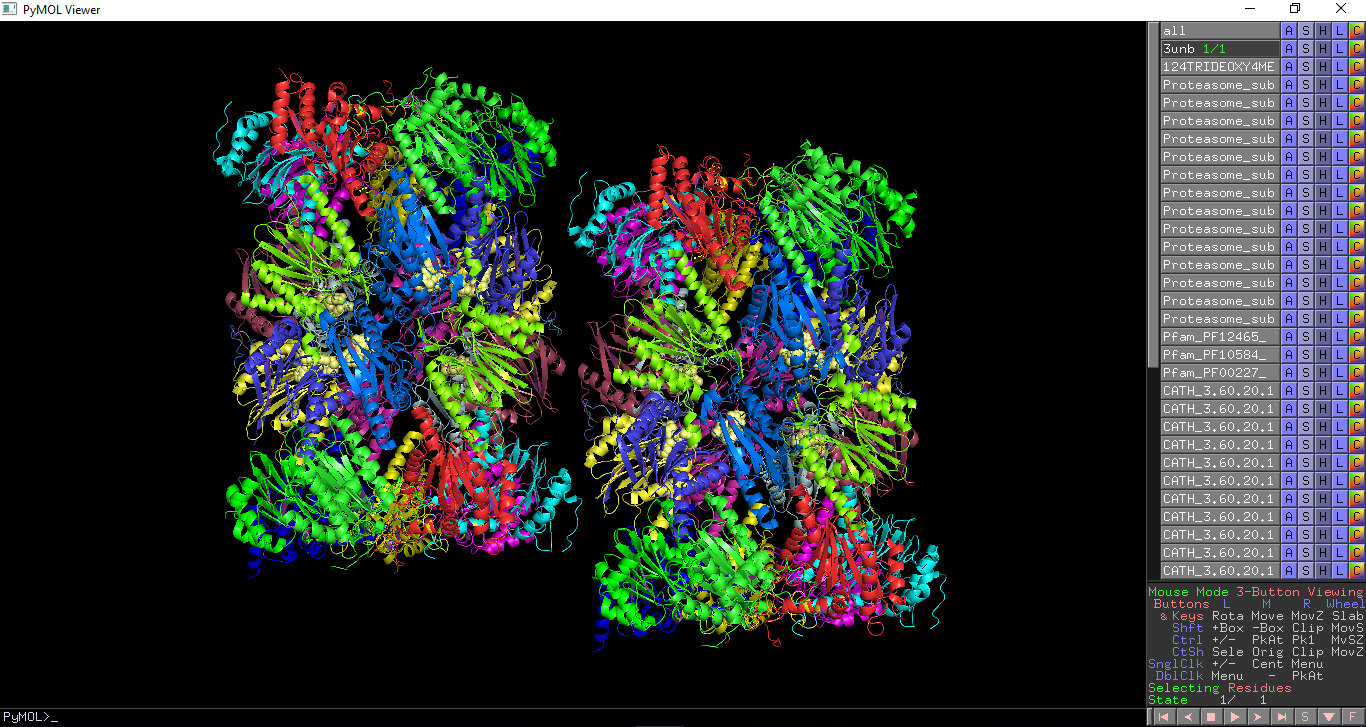
PDB Analysis - All
This displays chemically distinct molecules, domains which make up a PDB entry. It also displays the assemblies within a PDB entry.
Chemically distinct molecules which make up a PDB entry, including proteins, nucleic acids and ligands, are highlighted. Each molecule is given a unique colour and selected as a different object. Polymers (protein, DNA and RNA chains) are shown as cartoon or ribbon representation and ligands are shown as spheres. Pfam, SCOP and CATH domains within a PDB entry are highlighted. Each domain is coloured in a different colour and selected as a different object. This enables each domain to be turned on and off with PyMOL. Assemblies annotated within a PDB entry are highlighted. This requires pymol 1.76 or later.
All PDB annotations show in PDB entry 3unb
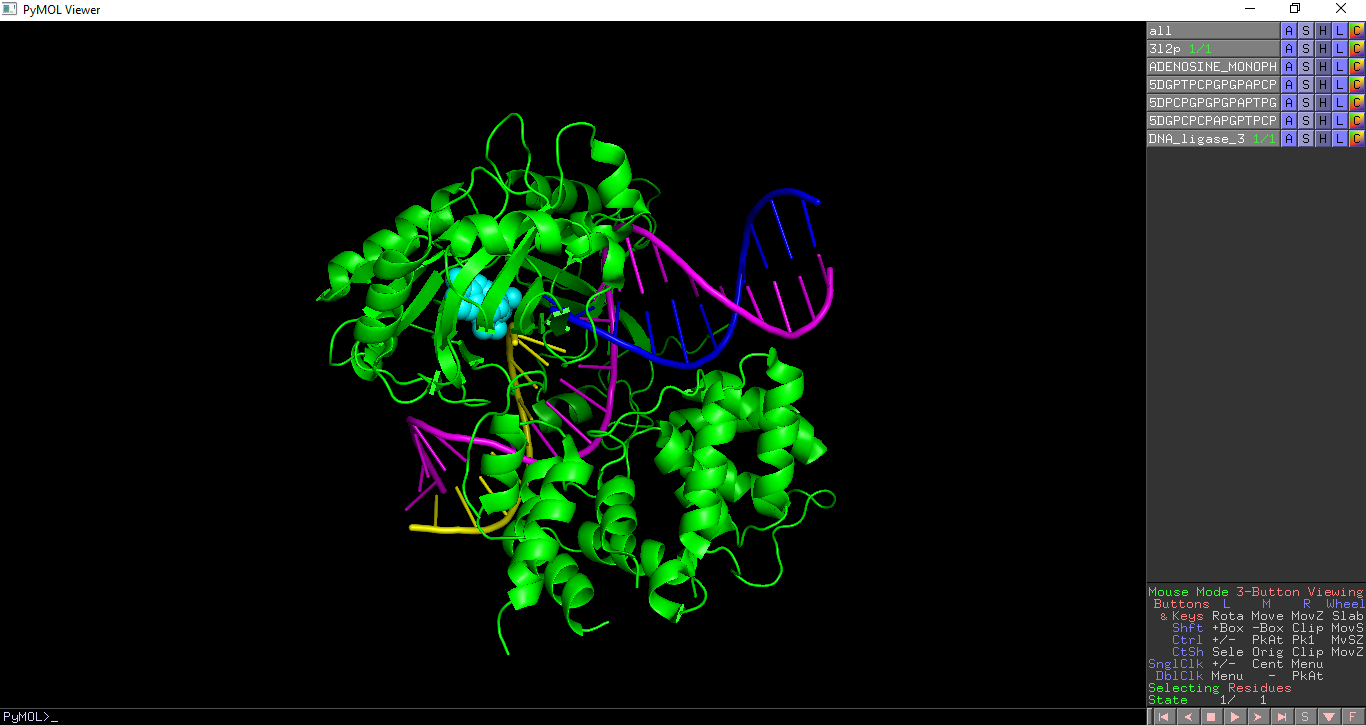
PDB Analysis - Molecules
This highlights the chemically distinct molecules which make up a PDB entry, including proteins, nucleic acids and ligands. Each molecule is given a unique colour and selected as a different object. Polymers (protein, DNA and RNA chains) are shown as cartoon or ribbon representation and ligands are shown as spheres.
In the below example the distinct molecules in PDB entry 3l2p are shown.
PDB Analysis - Domains
This highlights the Pfam, Rfam, SCOP and CATH domains within a PDB entry. Each domain is coloured in a different colour and selected as a different object. This enables each domain to be turned on and off with PyMOL. The Domains Plugin also highlights chemically distinct molecules and domains are overlaid on these molecules.
The CATH domains in PDB entry 3b43 are highlighted in the figure below.
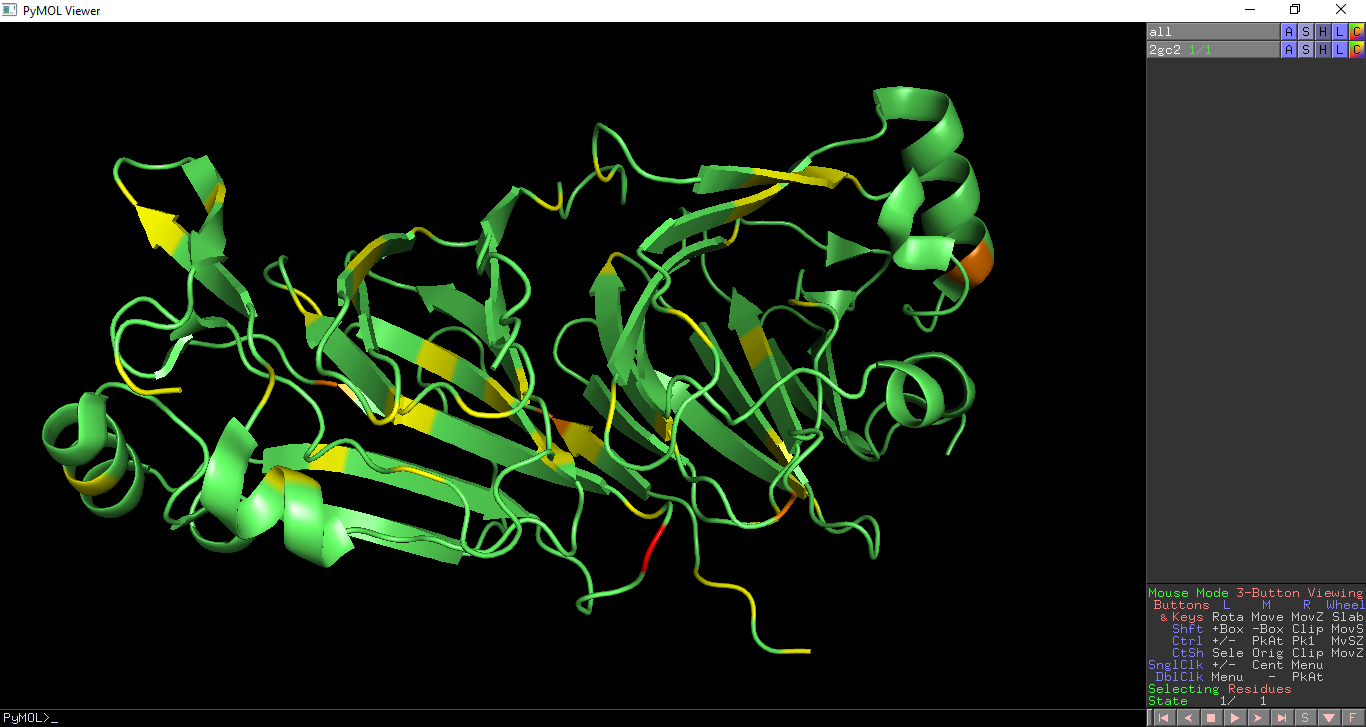
PDB Analysis - Validation
This overlays geometric validation on a PDB entry. Geometry outliers are coloured as per the PDB validation report: Residues with no geometric outliers are shown in green Residues with one geometric outliers are shown in yellow Residues with two geometric outliers are shown in orange Residues with three or more geometric outliers are shown in red.
Geometric validation is shown on PDB entry 2gc2
PDB Analysis - Assemblies
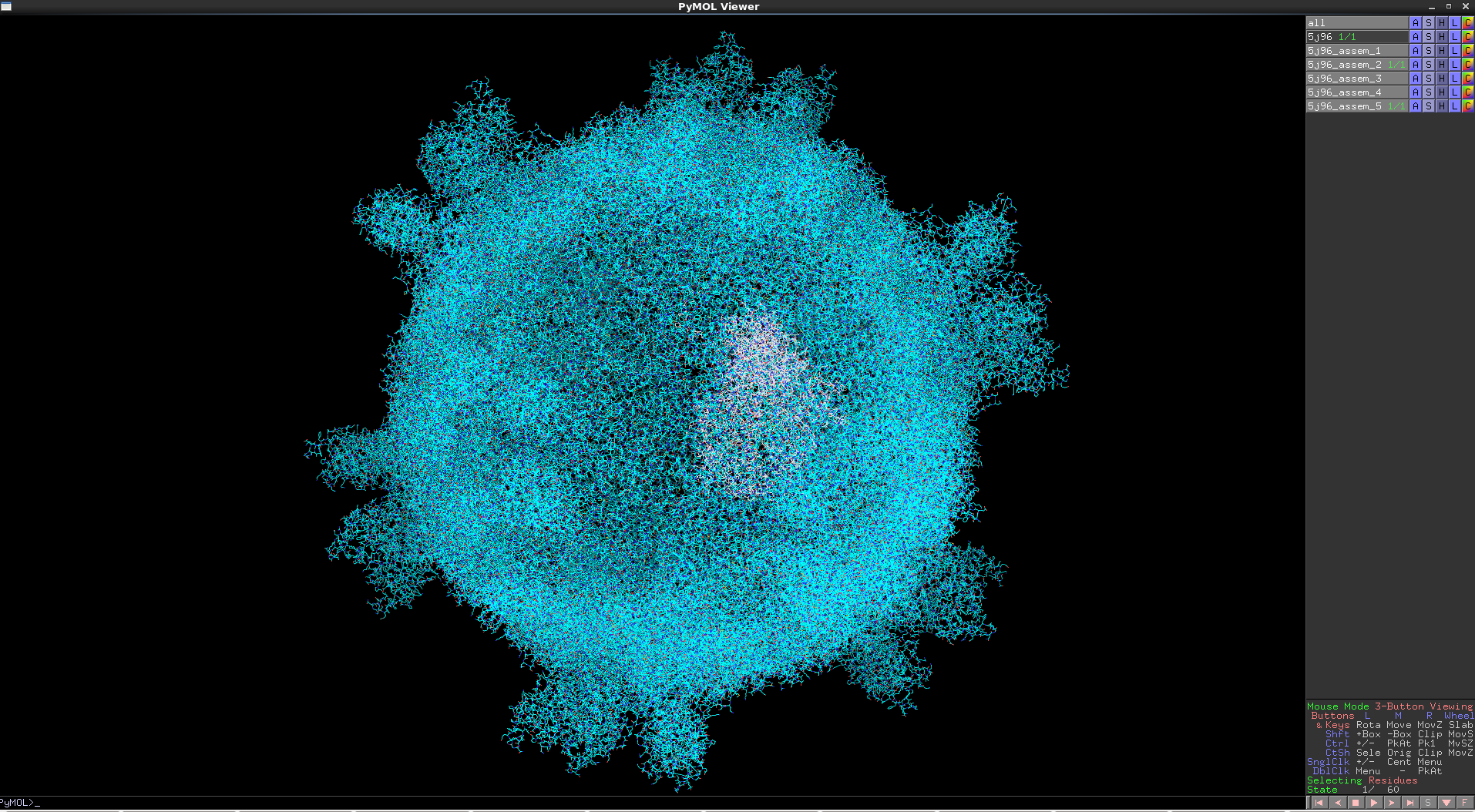
This highlights the assemblies within a PDB entry. This requires pymol 1.76 or later.
Assemblies are shown within PDB entry 5j96, with each assembly as an object in pymol and coloured in a distinct colour.
View assemblies for your own mmCIF file
To view annotated assemblies in your own mmCIF file - for example after PDB annotation - run pymol with the following command:
pymol -r PDB_plugin.py -- mmCIF_file=YOUR_MMCIF_FILE
where YOUR_MMCIF_FILE is the filename you want display the assemblies for.
Acknowledgements
This plugin was developed by PDBe
 who are a founding member of the wwPDB
who are a founding member of the wwPDB