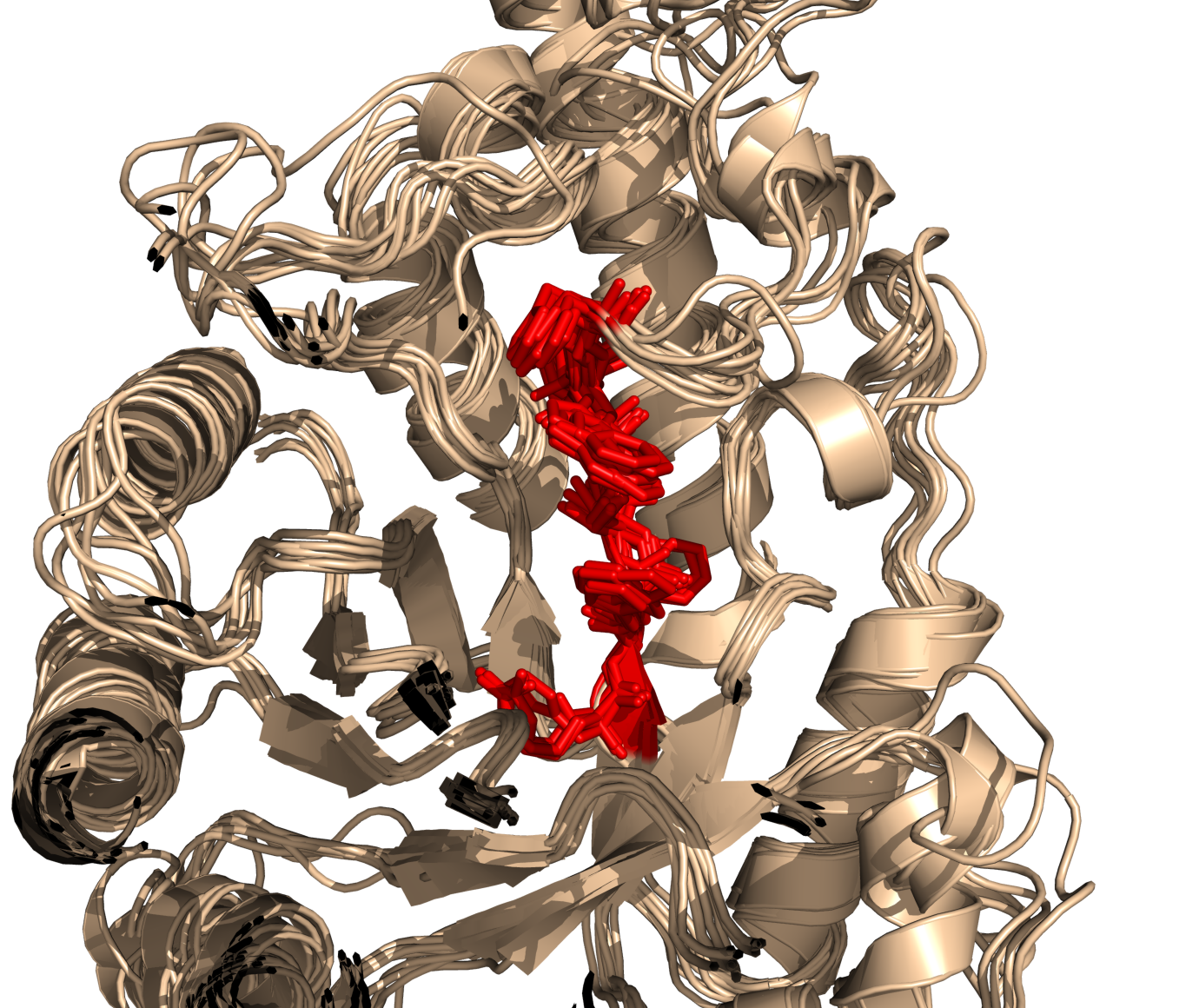
Anyone ever give you a protein and then say, find the sequence "FLVEW"? Well, this script will find any string or regular expression in a given object and return that selection for you. Here's an example,
reinitialize
import findseq
# fetch two sugar-binding PDB
fetch 1tvn, async=0
# Now, find FATEW in 1tvn, similarly
findseq FATEW, 1tvn
# lower-case works, too
findseq fatew, 1tvn
# how about a regular expression?
findseq F.*W, 1tvn
# Find the regular expression:
# ..H[TA]LVWH
# in the few proteins loaded.
# I then showed them as sticks and colored them to highlight matched AAs
for x in cmd.get_names(): findseq.findseq("..H[TA]LVWH", x, "sele_"+x, firstOnly=1)