News & Updates
| Warren
|
News about Warren DeLano's passing may be read on Warren's memorial page.
|
| PyMOLWiki
|
Reformatted Main Page. Some delays might occur from resizing images, but that should go away once the images are cached.
|
| PyMOLWiki
|
New — Search the PyMOLWiki via GoogleSearch
|
| PyMOL
|
PyMOL is known to work under Mac OS X 10+ (Snow Leopard) using the new Fink and incentive builds. See Installing PyMOL under Fink.
|
| PyMOL
|
PyMOL now has a Set command for basic settings, and a Set_bond command for bond-only settings.
|
| PyMOL
|
Space— new command to control PyMOL's usage of color spaces.
|
|
Did you know...
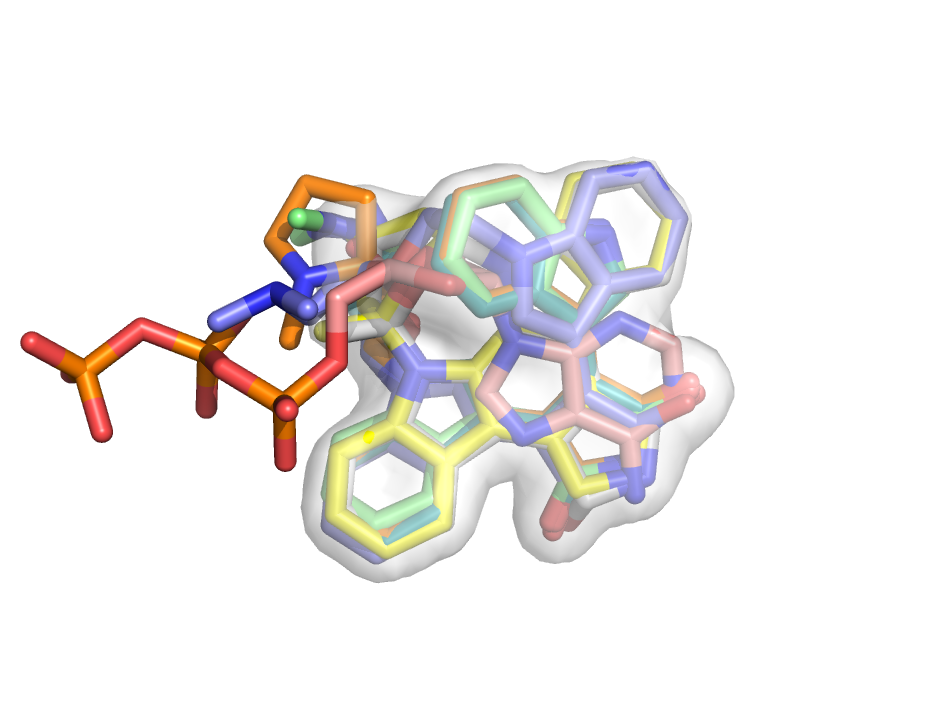 Map_set created this consensus volume representation of aligned ligands bound to kinases. See example below. Map_set provides a number of common operations on and between maps. For example, with Map_set you can add two maps together or create a consensus map from a series of maps or even take a difference map.
Usage
<source ..→
|
|
|
 A Random PyMOL-generated Cover. See Covers.
|

