COLORAMA
COLORAMA is a PyMOL plugin which allows to color objects using adjustable scale bars.
Program features
In RGB mode, each color R, G, B is represented by one scale bar which can be manually adjusted while the selected object is colored in real time. For convenience, it is as well possible to switch to the HSV color system.
Additionally, a color gradient with user-defined start- and end-colors can be created for the selected molecule.
Screenshot
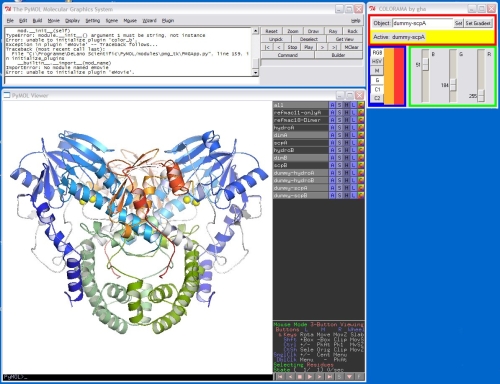
Usage
This plugin is included in the project Pymol-script-repo.
Manual install the plugin by copying the code below into an empty text file (e.g. "colorama.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the PLUGINS menu. COLORAMA has not been tested with PyMOL versions older than 1.0.
===Single ..→

