News & Updates
| New Plugin
|
Bondpack is a a collection of PyMOL plugins for easy visualization of atomic bonds.
|
| New Plugin
|
MOLE 2.0 is a new plugin for rapid analysis of biomacromolecular channels in PyMOL.
|
| 3D using Geforce
|
PyMOL can now be visualized in 3D using Nvidia GeForce video cards (series 400+) with 120Hz monitors and Nvidia 3D Vision, this was previously only possible with Quadro video cards.
|
| New Plugin
|
GROMACS_Plugin is a new plugin that ties together PyMOL and GROMACS.
|
| New Software
|
CMPyMOL is a software that interactively visualizes 2D contact maps of proteins in PyMOL.
|
| New Script
|
cgo_arrow draws an arrow between two picked atoms.
|
| Tips & Tricks
|
Instructions for generating movie PDFs using .mpg movies from PyMOL.
|
| Older News
|
See Older News.
|
|
Did you know...
= Overview =
Turn on or off the tracing of fog. Adding fog to the image assists in adding the feeling of depth.
See Also
depth_cue
|
|
|
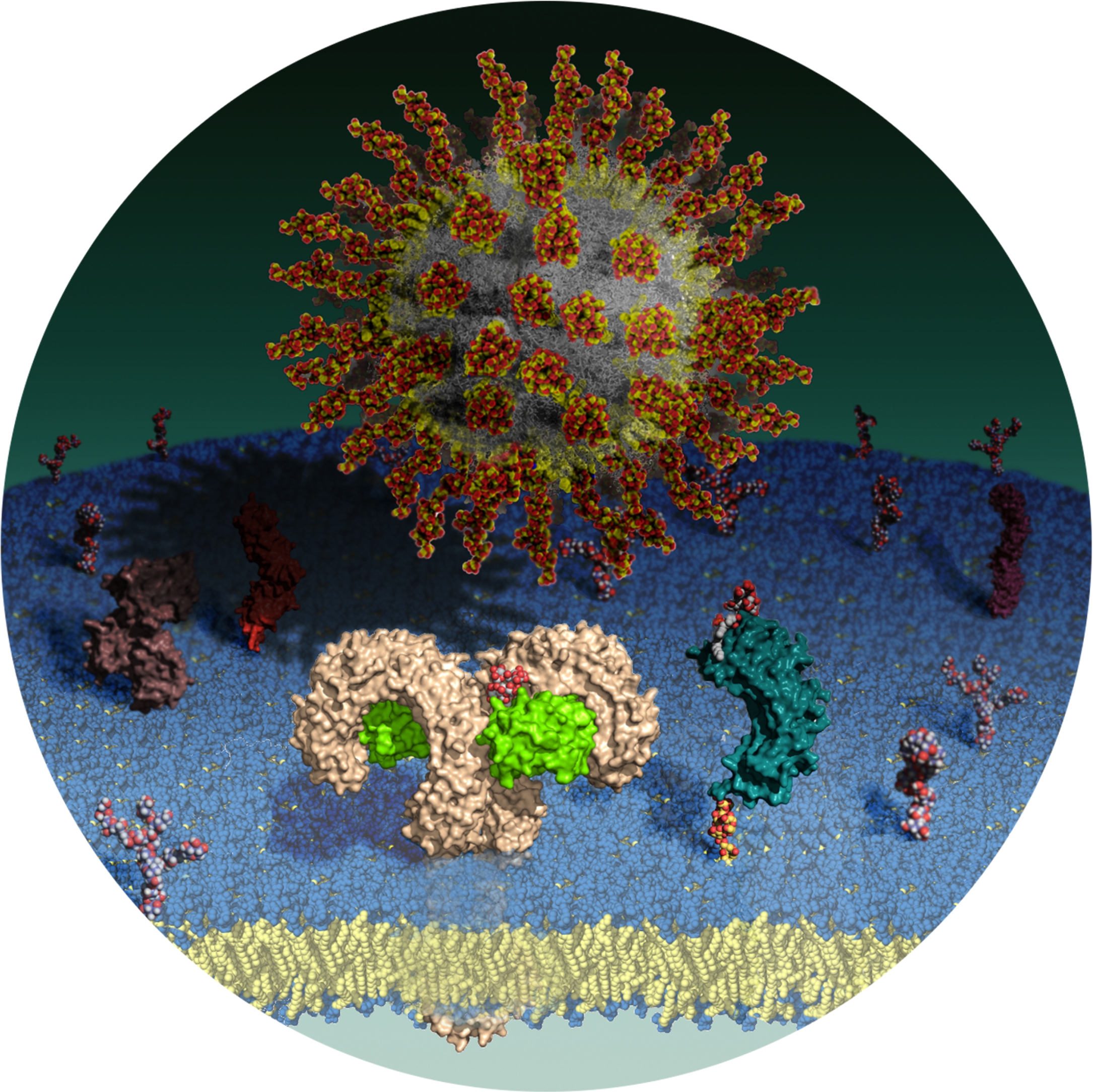 A Random PyMOL-generated Cover. See Covers.
|
