News & Updates
| New Script
|
spectrum_states colors states of multi-state object
|
| New Script
|
TMalign is a wrapper for the TMalign program
|
| Gallery Updates
|
The gallery has been updated to include a few new ideas and scripts for rendering
|
| New Script
|
save_settings can dump all changed settings to a file
|
| Tips & Tricks
|
Instructions for generating 3D PDFs using PyMOL.
|
| Wiki Update
|
Wiki has been updated. Please report any problems to the sysops.
|
| New Scripts
|
Create objects for each molecule or chain in selection with split_object and split_chains
|
| New Script
|
Rotkit: is a collection of usefull scripts to place your dye/molecule where you want. It includes a very handy, rotation around line, function.
|
| New Script
|
Forster-distance-calculator: Can be used as a pymol-python shortcut to calculate the Förster distance between dyes from different companies. Useful, if the user have pymol installed, but not python. This script is meant as a tool to finding the right dyes, when labelling suitable positions for the site-directed cysteine mutants. See DisplacementMap.
|
| New Script
|
propka: Fetches the pka values for your protein from the propka server. propka generates a pymol command file that make pka atoms, color and label them according to your protein. Inspection is made easy by grouping the pka atoms in the pymol menu.
|
| Older News
|
See Older News.
|
|
Did you know...
== Overview ==
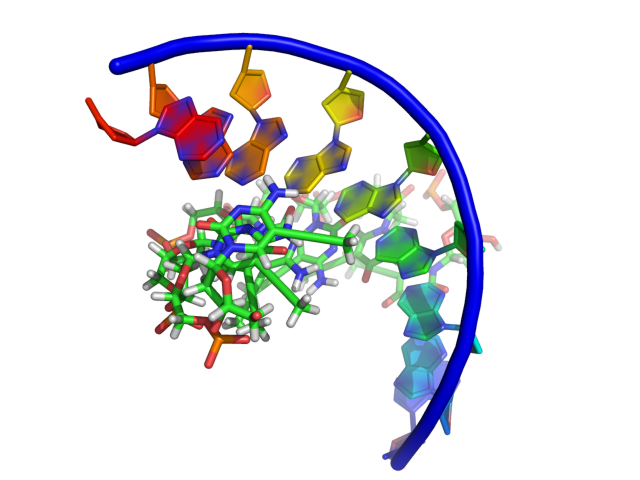
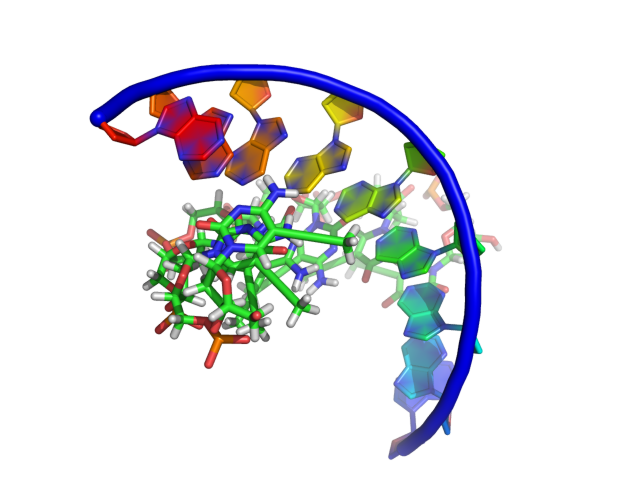
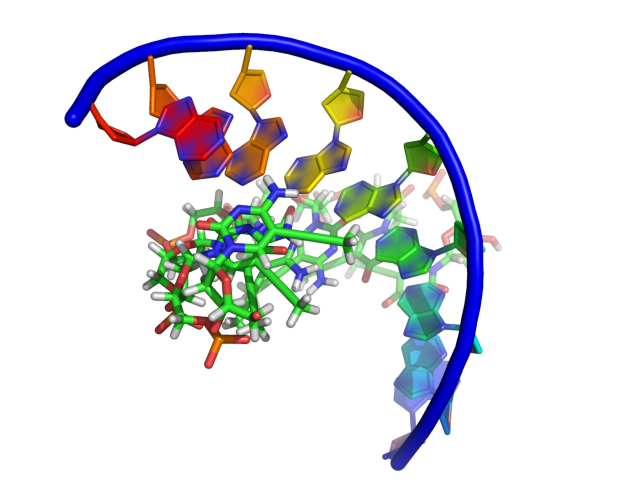
This setting modifies how PyMol draws the sugar bb of nucleic acids. Some differences between the settings are subtle, however PyMol gives you the ability to get the representation you want. Click on each images for a closer look.
Examples
Syntax
<source lang="python">
set cartoon_nucleic_acid_mode, 1 # try different ..→
|
|
|
 A Random PyMOL-generated Cover. See Covers.
|