Elbow angle: Difference between revisions
Jump to navigation
Jump to search
Jaredsampson (talk | contribs) No edit summary |
Jaredsampson (talk | contribs) (→Examples: removed gallery dimensions) |
||
| Line 21: | Line 21: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
<gallery | <gallery> | ||
Image:elbow_angle_3ghe.png|Fab fragment 3ghe shown with draw=1. | Image:elbow_angle_3ghe.png|Fab fragment 3ghe shown with draw=1. | ||
Image:elbow_angle_2.png|5 PDB examples from Stanfield, et al., JMB 2006, shown in the same orientation as in Figure 1 of that paper. | Image:elbow_angle_2.png|5 PDB examples from Stanfield, et al., JMB 2006, shown in the same orientation as in Figure 1 of that paper. | ||
Revision as of 13:42, 8 June 2012
| Type | Python Script |
|---|---|
| Download | elbow_angle.py |
| Author(s) | Jared Sampson |
| License | GPLv3 |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
This script allows you to calculate the elbow angle of an antibody Fab fragment object and optionally draw a graphical representation of the vectors used to calculate the elbow angle.
Examples
# load an antibody Fab from the PDB
fetch 3ghe, async=0
# get a nice orientation
orient
# calculate the elbow angle and draw the vectors
elbow_angle 3ghe, draw=1
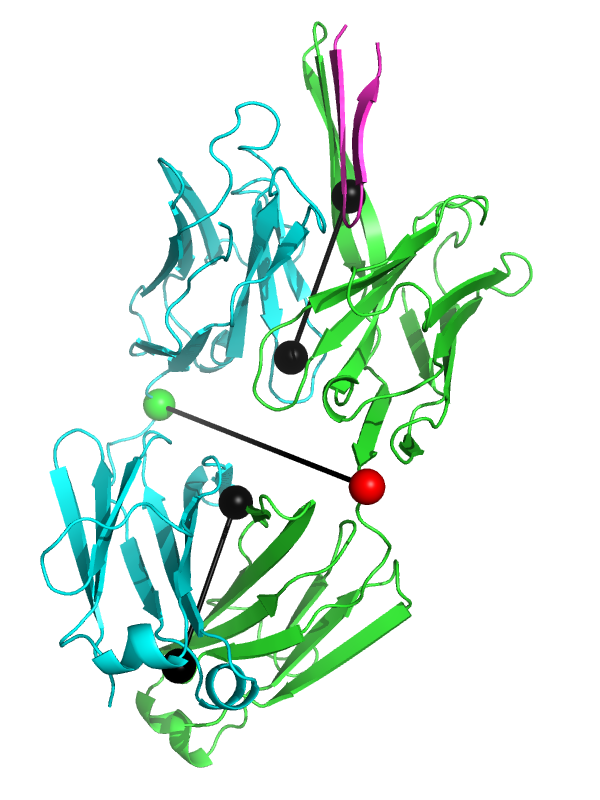
- Elbow angle 2.png
5 PDB examples from Stanfield, et al., JMB 2006, shown in the same orientation as in Figure 1 of that paper.
The black "dumbbells" pass through the centers of mass of the combined variable and constant domains, respectively. The green and red dumbbell denotes the residues used to split the variable and constant domains, with a green ball for the light chain, and a red ball for the heavy chain.