Alter: Difference between revisions
Jump to navigation
Jump to search
| Line 61: | Line 61: | ||
</gallery> | </gallery> | ||
<source lang="python"> | <source lang="python"> | ||
# create a pseudoatom at the origin; we will | |||
# measure the distance from this point | |||
pseudoatom pOrig, pos=(0,0,0), label=origin | |||
# fetch and build the capsid | # fetch and build the capsid | ||
| Line 71: | Line 76: | ||
as surface | as surface | ||
# create a new color ramp, measuring the distance | # create a new color ramp, measuring the distance | ||
Revision as of 13:10, 29 July 2011
alter changes one or more atomic properties over a selection using the python evaluator with a separate name space for each atom. The symbols defined in the name space, which are are explained in Iterate are:
name, resn, resi, chain, alt, elem, q, b, segi, type (ATOM,HETATM), partial_charge, formal_charge, text_type, numeric_type, ID, vdw
All strings must be explicitly quoted. This operation typically takes several seconds per thousand atoms altered.
WARNING: You should always issue a sort command on an object after modifying any property which might affect canonical atom ordering (names, chains, etc.). Failure to do so will confound subsequent "create" and "byres" operations.
USAGE
alter (selection),expression
EXAMPLES
Change chain label and residue index
alter (chain A),chain='B'
alter (all),resi=str(int(resi)+100)
sort
Change van der Waals radius of a given atom
alter (name P), vdw=1.90
Note that is if dots, spheres, mesh or surface representation is used. You have to refresh the view with
rebuild
Renumber the amino acids in a protein, so that it starts from 0 instead of its offset as defined in the structure file
# The first residue in the structure file for 1cll is 4. To change this to 0, maybe to match scripts
# outputted from other programs, just remove the offset of 4 from each atom
alter 1cll, resi=str(int(resi)-4)
# refresh (turn on seq_view to see what this command does).
sort
Change the b values of all atoms to the distance of the atoms to a reference point
# reference point
x0,y0,z0=[1,2,3]
# calculate distance values between the reference point and all the atoms
alldist = []
iterate_state 1, yourstruc, alldist.append(((x-x0)**2.0+(y-y0)**2.0+(z-z0)**2.0)**0.5)
# assign distance values to b-factors
di = iter(alldist)
alter yourstruc, b=di.next()
# visualize the distances
spectrum b, rainbow, yourstruc
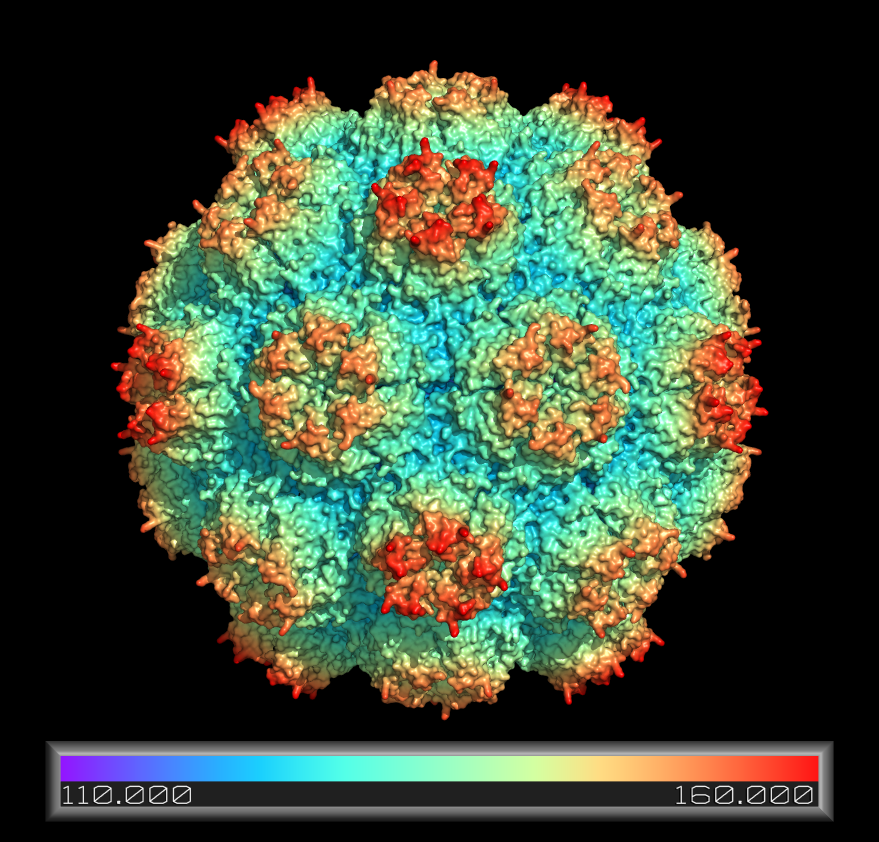
Coloring a Viral Capsid by Distance from Core
# create a pseudoatom at the origin; we will
# measure the distance from this point
pseudoatom pOrig, pos=(0,0,0), label=origin
# fetch and build the capsid
fetch 2xpj, async=0, type=pdb1
split_states 2xpj
delete 2xpj
# show all 60 subunits it as a surface
# this will take a few minutes to calculate
as surface
# create a new color ramp, measuring the distance
# from pOrig to 1hug, colored as rainbow
ramp_new proximityRamp, pOrig, selection=(2xpj*), range=[110,160], color=rainbow
# set the surface color to the ramp coloring
set surface_color, proximityRamp, (2xpj*)
# some older PyMOLs need this recoloring/rebuilding
recolor