FindSurfaceResidues: Difference between revisions
Jump to navigation
Jump to search
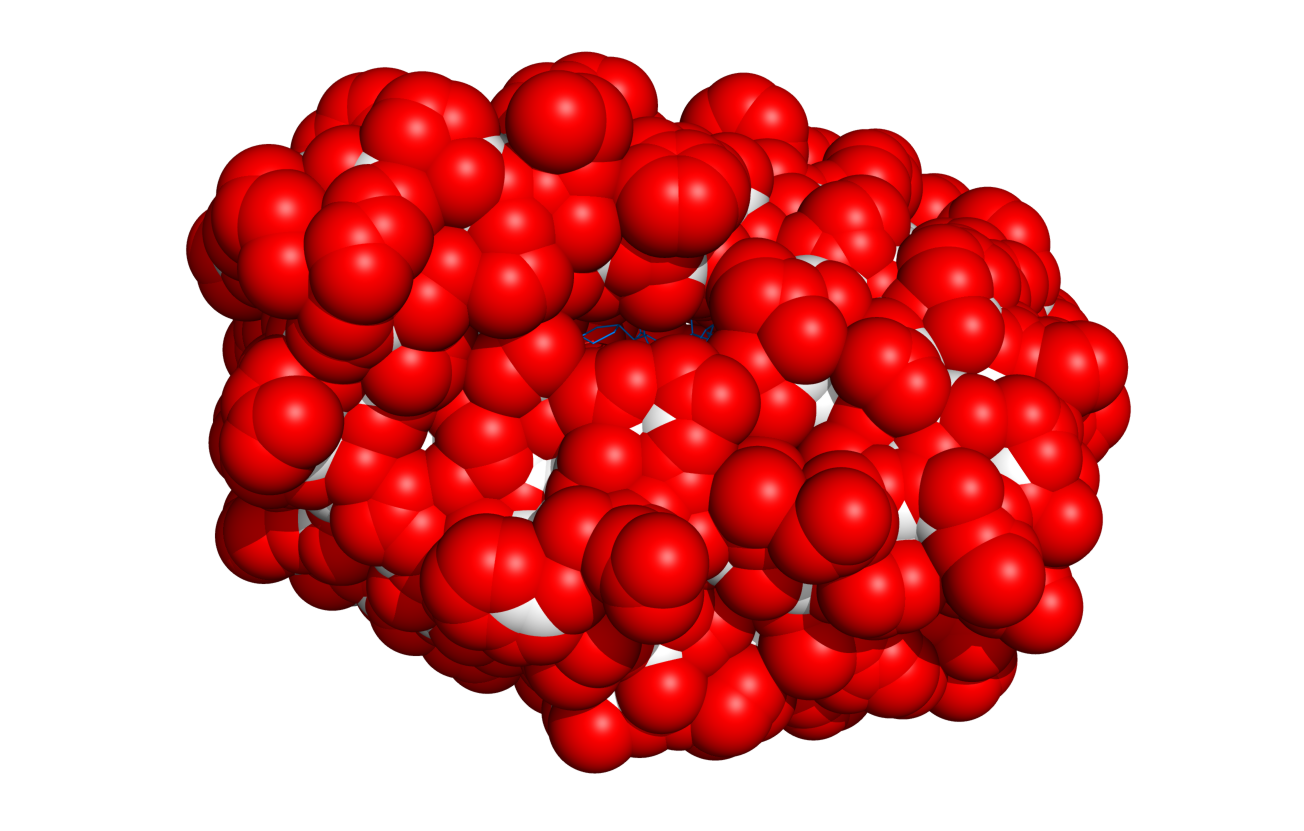
(Created page with 'thumb|right|400px|Result of $TUT/1hpv.pdb at 2.5 Ang cutoff. = Overview = This script will select (and color if requested) surface residues on an object...') |
|||
| Line 38: | Line 38: | ||
# watch how the visualization changes: | # watch how the visualization changes: | ||
findSurfaceResidues doShow=True, cutoff=0.5 | |||
findSurfaceResidues doShow=True, cutoff=1.0 | |||
findSurfaceResidues doShow=True, cutoff=1.5 | |||
findSurfaceResidues doShow=True, cutoff=2.0 | |||
findSurfaceResidues doShow=True, cutoff=2.5 | |||
findSurfaceResidues doShow=True, cutoff=3.0 | |||
</source> | </source> | ||
Revision as of 11:18, 17 June 2009
Overview
This script will select (and color if requested) surface residues on an object or selection. See the options below.
Each time, the script will create a new selection called, 'exposedXYZ' where XYZ is some random number. This is done so that no other selections/objects are overwritten.
Usage
findSurfaceResidues [objSel=(all)[, cutoff=2.5[, doShow=False[, verbose=False ]]]]
The parameters are:
objSel
- The object or selection for which to find exposed residues;
- DEFAULT = (all)
cutoff
- The cutoff in square Angstroms that defines exposed or not. Those residues with > cutoff Ang^2 exposed will be considered exposed;
- DEFAULT = 2.5 Ang^2
doShow
- Change the visualization to highlight the exposed residues vs interior
- DEFAULT = False/Blank
verbose
- Level of verbosity.
- DEFAULT = False/Blank
Examples
# make sure you download and run the code below, before trying these examples.
load $TUT/1hpv.pdb
findSurfaceResidues
# now show the exposed
findSurface residues doShow=True
# watch how the visualization changes:
findSurfaceResidues doShow=True, cutoff=0.5
findSurfaceResidues doShow=True, cutoff=1.0
findSurfaceResidues doShow=True, cutoff=1.5
findSurfaceResidues doShow=True, cutoff=2.0
findSurfaceResidues doShow=True, cutoff=2.5
findSurfaceResidues doShow=True, cutoff=3.0
The Code
# -*- coding: utf-8 -*-
import pymol
from pymol import cmd
import random
def findSurfaceResidues(objSel="(all)", cutoff=2.5, doShow=False, verbose=False):
"""
findSurfaceResidues
finds those residues on the surface of a protein
that have at least 'cutoff' exposed A**2 surface area.
PARAMS
objSel (string)
the object or selection in which to find
exposed residues
DEFAULT: (all)
cutoff (float)
your cutoff of what is exposed or not.
DEFAULT: 2.5 Ang**2
asSel (boolean)
make a selection out of the residues found
RETURNS
(list: (chain, resv ) )
A Python list of residue numbers corresponding
to those residues w/more exposure than the cutoff.
"""
tmpObj="__tmp"
cmd.create( tmpObj, objSel + " and polymer");
if verbose!=False:
print "WARNING: I'm setting dot_solvent. You may not care for this."
cmd.set("dot_solvent");
cmd.get_area(selection=tmpObj, load_b=1)
# threshold on what one considers an "exposed" atom (in A**2):
cmd.remove( tmpObj + " and b < " + str(cutoff) )
stored.tmp_dict = {}
cmd.iterate(tmpObj, "stored.tmp_dict[(chain,resv)]=1")
exposed = stored.tmp_dict.keys()
exposed.sort()
selName = "exposed_" + str(random.randint(0,10000))
if verbose!=False:
print "Exposed residues are selected in: " + selName
cmd.select(selName, objSel + " in " + tmpObj )
if doShow!=False:
cmd.show_as("spheres", objSel + " and poly")
cmd.color("white", objSel)
cmd.color("red", selName)
cmd.delete(tmpObj)
return exposed
cmd.extend("findSurfaceResidues", findSurfaceResidues)