Symexp: Difference between revisions
mNo edit summary |
(naming scheme) |
||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Symexp]] is used to reconstruct neighboring asymmetric units from the crystallographic experiment that produced the given structure. This is assuming the use of a [http://www.rcsb.org/pdb/home/home.do PDB] file or equivalent that contains enough information to reproduce the lattice. | [[Symexp]] is used to reconstruct neighboring asymmetric units from the crystallographic experiment that produced the given structure. This is assuming the use of a [http://www.rcsb.org/pdb/home/home.do PDB] file or equivalent that contains enough information ([http://www.wwpdb.org/documentation/file-format-content/format33/sect8.html#CRYST1 CRYST1 record]) to reproduce the lattice. | ||
[[Symexp]] creates all symmetry related objects for the specified object that occurs within a cutoff about an atom selection. The new objects are labeled using the prefix provided along with their crystallographic symmetry operation and translation. | [[Symexp]] creates all symmetry related objects for the specified object that occurs within a cutoff about an atom selection. The new objects are labeled using the prefix provided along with their crystallographic symmetry operation and translation. | ||
= USAGE = | == USAGE == | ||
<source lang="python"> | <source lang="python"> | ||
# Expand the ''object'' around its ''selection'' by cutoff Angstroms and | # Expand the ''object'' around its ''selection'' by cutoff Angstroms and | ||
# prefix the new objects withs ''prefix''. | # prefix the new objects withs ''prefix''. | ||
symexp prefix, object, selection[, | symexp prefix, object, selection, cutoff [, segi] | ||
</source> | </source> | ||
| Line 14: | Line 14: | ||
symexp name_for_new_objects,asymmetric_name,(asymmetric_name),distance | symexp name_for_new_objects,asymmetric_name,(asymmetric_name),distance | ||
</source> | </source> | ||
== ARGUMENTS == | |||
= EXAMPLE = | * '''prefix''' = string: name prefix for new objects | ||
* '''object''' = string: name of the object that you wish to reproduce neighboring crystal partners for; the source of the symmetry operators | |||
* '''selection''' = string: atom selection to measure cutoff distance from | |||
* '''cutoff''' = float: create all symmetry mates that are within "cutoff" distance from selection (but no more than +/-1 unit cells) | |||
* '''segi''' = 0/1: if segi=1 then assign to each symmetry mate a unique 4-character segment identifier {default: 0} | |||
== EXAMPLE == | |||
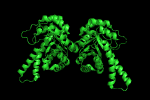
load any .pdb file into PyMOL (here we use 1GVF).<br> | load any .pdb file into PyMOL (here we use 1GVF).<br> | ||
fetch 1GVF | |||
[[Image:1GVF_assym.png]] | [[Image:1GVF_assym.png]] | ||
| Line 49: | Line 50: | ||
PyMOL is known to exit dramatically (crash) if you provide a scene that is too large or complex. This is a result of the low-level ''malloc'' function failing. See [[:Category:Performance]] for workarounds. | PyMOL is known to exit dramatically (crash) if you provide a scene that is too large or complex. This is a result of the low-level ''malloc'' function failing. See [[:Category:Performance]] for workarounds. | ||
= See Also = | == Naming Scheme == | ||
The created objects have the following naming scheme: | |||
<prefix>AAXXYYZZ | |||
* AA: zero-based [https://sourceforge.net/p/pymol/code/HEAD/tree/trunk/pymol/modules/pymol/xray.py#l95 symmetry operator] index, e.g. 00 always corresponds to "x,y,z". A space group like "P 21 21 21" which has 4 symmetry operators will count up to 03 | |||
* XX: -1, 0, or 1, this is the unit cell offset from the center of the selection along the x-axis. PyMOL never creates more than 3 unit cells along each axis | |||
* YY: analogous to XX, but along the y-axis | |||
* ZZ: analogous to XX, but along the z-axis | |||
== See Also == | |||
* [http://pdbbeta.rcsb.org/robohelp_f/data_download/biological_unit/pdb_and_mmcif_files_.htm PDB Symmetry Info] | * [http://pdbbeta.rcsb.org/robohelp_f/data_download/biological_unit/pdb_and_mmcif_files_.htm PDB Symmetry Info] | ||
* [[SuperSym]] | * [[SuperSym]] | ||
* [[Supercell]] | |||
* From within PyMOL, ''help symexp'' and ''symexp ?''. | * From within PyMOL, ''help symexp'' and ''symexp ?''. | ||
[[Category:Commands|Symexp]] | [[Category:Commands|Symexp]] | ||
Latest revision as of 10:48, 20 April 2018
Symexp is used to reconstruct neighboring asymmetric units from the crystallographic experiment that produced the given structure. This is assuming the use of a PDB file or equivalent that contains enough information (CRYST1 record) to reproduce the lattice.
Symexp creates all symmetry related objects for the specified object that occurs within a cutoff about an atom selection. The new objects are labeled using the prefix provided along with their crystallographic symmetry operation and translation.
USAGE
# Expand the ''object'' around its ''selection'' by cutoff Angstroms and
# prefix the new objects withs ''prefix''.
symexp prefix, object, selection, cutoff [, segi]
For one protein:
symexp name_for_new_objects,asymmetric_name,(asymmetric_name),distance
ARGUMENTS
- prefix = string: name prefix for new objects
- object = string: name of the object that you wish to reproduce neighboring crystal partners for; the source of the symmetry operators
- selection = string: atom selection to measure cutoff distance from
- cutoff = float: create all symmetry mates that are within "cutoff" distance from selection (but no more than +/-1 unit cells)
- segi = 0/1: if segi=1 then assign to each symmetry mate a unique 4-character segment identifier {default: 0}
EXAMPLE
load any .pdb file into PyMOL (here we use 1GVF).
fetch 1GVF
At the PyMOL command prompt type the following:
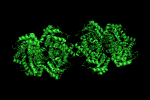
symexp sym,1GVF,(1GVF),1
produces three new objects. We now have four objects corresponding to two biologic units (the functional protein in a cell).
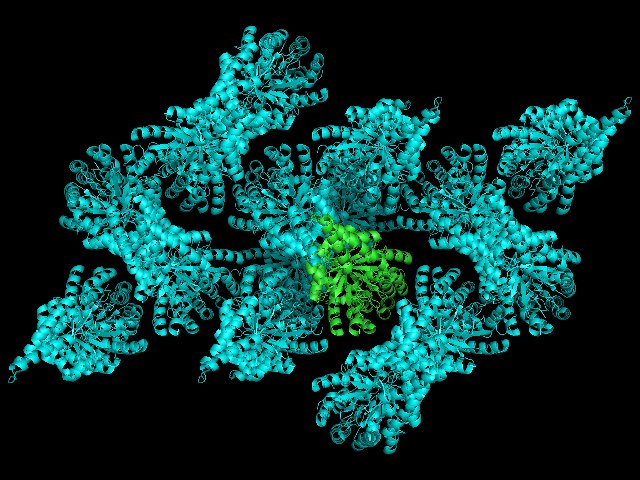
symexp sym,1GVF,(1GVF),5
If we color all of the sym* cyan we will produce the following:
As you can see, we can begin to understand the crystal environment of our asymmetric unit. Increasing distance will reveal more of the crystal lattice, but will place in increasing demand on your computer's rendering ability.
PyMOL is known to exit dramatically (crash) if you provide a scene that is too large or complex. This is a result of the low-level malloc function failing. See Category:Performance for workarounds.
Naming Scheme
The created objects have the following naming scheme:
<prefix>AAXXYYZZ
- AA: zero-based symmetry operator index, e.g. 00 always corresponds to "x,y,z". A space group like "P 21 21 21" which has 4 symmetry operators will count up to 03
- XX: -1, 0, or 1, this is the unit cell offset from the center of the selection along the x-axis. PyMOL never creates more than 3 unit cells along each axis
- YY: analogous to XX, but along the y-axis
- ZZ: analogous to XX, but along the z-axis
See Also
- PDB Symmetry Info
- SuperSym
- Supercell
- From within PyMOL, help symexp and symexp ?.