Huge surfaces: Difference between revisions
Appearance
New page: = Overview = PyMOL can render '''very large''' surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you... |
CA-only |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
PyMOL can render '''very large''' (huge) surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you would for a smaller molecule. However, if you need to represent a molecule (or object) with a few million atoms, this may be your option of choice. | |||
PyMOL can render '''very large''' surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you would for a smaller molecule. However, if you need to represent a molecule (or object) with a few million atoms, this may be your option of choice. | |||
= Example = | = Example = | ||
| Line 10: | Line 9: | ||
</gallery> | </gallery> | ||
= | == Strategy 1: Low resolution Gaussian Map == | ||
<source lang="python"> | <source lang="python"> | ||
# load a whopping big PDB | # load a whopping big PDB | ||
fetch 1aon, struct | fetch 1aon, struct, async=0 | ||
# create a color spectrum over the object | # create a color spectrum over the object | ||
spectrum count, selection=struct | spectrum count, selection=struct | ||
# === now create a pseudo-fcalc map (a 3D volumetric scalar field) === | # === now create a pseudo-fcalc map (a 3D volumetric scalar field) === | ||
# set the B-factors nice and high for smoothness | # set the B-factors nice and high for smoothness | ||
set gaussian_b_floor, 40 | |||
# ~10 A map resolution | # ~10 A map resolution | ||
| Line 37: | Line 32: | ||
# now color the map based on the underlying protein | # now color the map based on the underlying protein | ||
ramp_new ramp, struct, [0 | ramp_new ramp, struct, [0,10], [atomic, atomic] | ||
color ramp, surf | color ramp, surf | ||
| Line 44: | Line 39: | ||
disable ramp | disable ramp | ||
</source> | </source> | ||
== Strategy 2: CA-only model with oversized radii == | |||
<source lang="python"> | |||
# load a whopping big PDB | |||
fetch 1aon, struct, async=0 | |||
# reduce to CA atoms only | |||
remove struct & not guide | |||
# create a color spectrum over the object | |||
spectrum count, selection=struct | |||
# increase VDW radii to compensate volume of missing non-CA atoms | |||
alter struct, vdw=4 | |||
# increase VDW radius of solvent | |||
set solvent_radius, 4, struct | |||
# show molecular surface | |||
as surface | |||
</source> | |||
[[Category:Performance]] | |||
Latest revision as of 17:55, 16 March 2018
PyMOL can render very large (huge) surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you would for a smaller molecule. However, if you need to represent a molecule (or object) with a few million atoms, this may be your option of choice.
Example
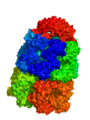
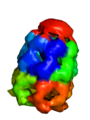
[edit]The output from the example below is shown below. The PDB 1AON has nearly 60,000 atoms. This isn't the largest PDB, but it's a good example.
Strategy 1: Low resolution Gaussian Map
[edit]# load a whopping big PDB
fetch 1aon, struct, async=0
# create a color spectrum over the object
spectrum count, selection=struct
# === now create a pseudo-fcalc map (a 3D volumetric scalar field) ===
# set the B-factors nice and high for smoothness
set gaussian_b_floor, 40
# ~10 A map resolution
set gaussian_resolution, 10
# ~10 A map spacing with a 10 A surrounding buffer
# (you may need to vary this)
map_new map, gaussian, 10, struct, 10
# create a surface from the map
isosurface surf, map, 1.0
# now color the map based on the underlying protein
ramp_new ramp, struct, [0,10], [atomic, atomic]
color ramp, surf
# hide the ramp
disable ramp
Strategy 2: CA-only model with oversized radii
[edit]# load a whopping big PDB
fetch 1aon, struct, async=0
# reduce to CA atoms only
remove struct & not guide
# create a color spectrum over the object
spectrum count, selection=struct
# increase VDW radii to compensate volume of missing non-CA atoms
alter struct, vdw=4
# increase VDW radius of solvent
set solvent_radius, 4, struct
# show molecular surface
as surface