Difference between revisions of "Valence"
Jump to navigation
Jump to search
m (repaired misspelled unbond link) |
|||
| Line 42: | Line 42: | ||
| − | =SEE ALSO= | + | ==Automatic editing of bonds== |
| + | [[Format_bonds]] is a script that automatically formats valence in amino acids. | ||
| + | |||
| + | |||
| + | ==SEE ALSO== | ||
[[Bond]], [[Unbond]] | [[Bond]], [[Unbond]] | ||
[[Category:Settings|Valence]] | [[Category:Settings|Valence]] | ||
Revision as of 16:42, 9 June 2014
Overview
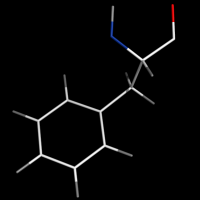
Turning on valence will enable the display of double bonds.
Toggling valence_mode alters the positioning of double bonds (for representation as Lines)
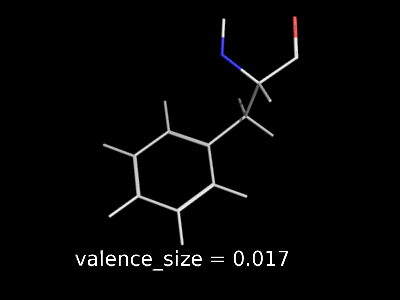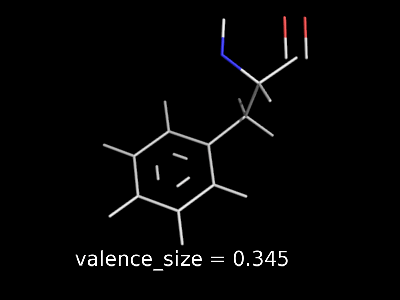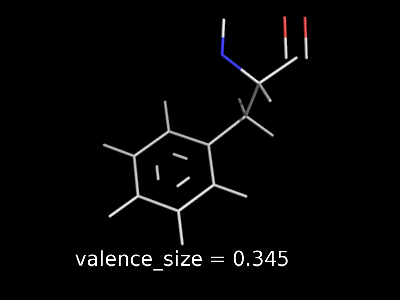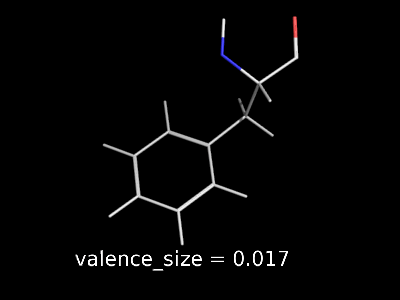
valence_size alters the distance of double bonds.
Note that bonds can be edited to be delocalized using Unbond and Bond.
Examples
set valence, 1
#delocalized bonds
#(edited: see Bond)
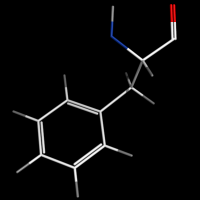
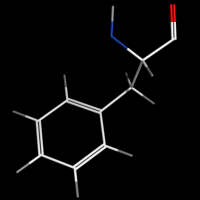
valence_size alters the distance of double bonds, but behaves slightly different depending on valence_mode
| valence_size with valence_mode 1 inside |
valence_size with valence_mode 0 centered |
|---|---|
 |
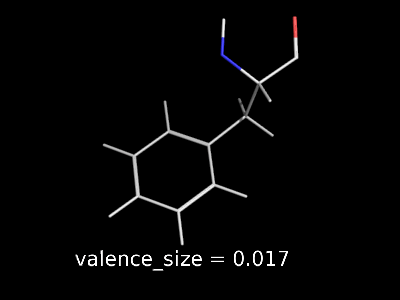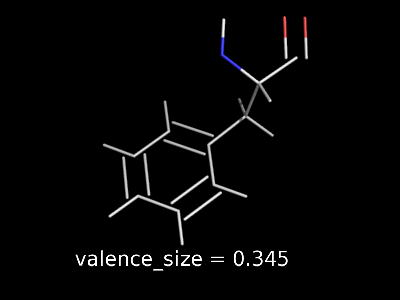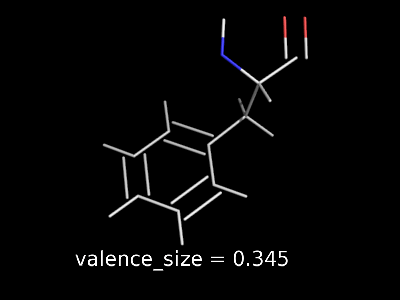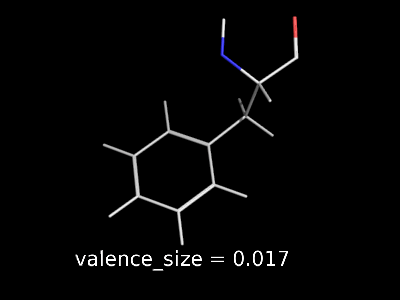
|
Syntax
set valence, 0 # off
set valence, 1 # on
set valence_mode, 0 # centered
set valence_mode, 1 # inside
set valence_size, 0.1 # default: 0.06 # range 0 - ~0.5
# Editing bonds:
# In editing mode: select the bond using Ctrl-right-click, then enter:
unbond pk1,pk2
bond pk1,pk2,4
# 1: single bond, 2: double bond, 3:triple bond, 4:delocalized
Automatic editing of bonds
Format_bonds is a script that automatically formats valence in amino acids.